ORIGINAL ARTICLE |
https://doi.org/10.5005/jp-journals-10071-23916 |
Increasing Trend of Vancomycin-resistant Enterococci Bacteremia in a Tertiary Care Hospital of South India: A Three-year Prospective Study
1–3,5,6Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India
4Yashoda Hospital, Hyderabad, Telangana, India
Corresponding Author: Apurba S Sastry, Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India, Phone: +91 9444327314, e-mail: drapurbasastry@gmail.com
How to cite this article: Sivaradjy M, Gunalan A, Priyadarshi K, Madigubba H, Rajshekar D, Sastry AS. Increasing Trend of Vancomycin-resistant Enterococci Bacteremia in a Tertiary Care Hospital of South India: A Three-year Prospective Study. Indian J Crit Care Med 2021;25(8):881–885.
Source of support: Nil
Conflict of interest: None
ABSTRACT
Introduction: Vancomycin-resistant enterococci (VRE) are emerging as an important multidrug-resistant pathogen causing nosocomial infections, predominantly bacteremia and urinary tract infections. VRE bacteremia has caused a significant increase in the duration of the hospital stay and mortality and had caused high public health threat due to limited treatment options.
Materials and methods: Between October 2017 and September 2020, all consecutive patients with culture-proven bloodstream infection with Enterococcus species, isolated for the first time, were included in the study. A total of 427 Enterococcus species were identified, and antimicrobial susceptibility tests were performed and interpreted using Clinical and Laboratory Standard Institute guidelines.
Results: Of the total 427 Enterococcus species isolated, 63 (45.6%) were VRE. Among them, 51/63 (81%) were Enterococcus faecium (E. faecium) and 5/63 (8%) were Enterococcus faecalis. There was an increased trend of VRE rate in the bloodstream infections of 6.12% (2018), 13.2% (2019), and 19.2% (2020). The majority of the VRE patients [43/63 (68%)] were admitted to the intensive care units (ICUs). Vancomycin A (VanA) is the most common phenotype isolated from 51/63(81%) patients.
Conclusion: This increasing trend of VRE bacteremia is a red alert to the clinicians and the infection control practitioners, so that strict antibiotic policies and proper adherence to the infection control practices can be initiated to reduce the VRE rate.
Keywords: Nosocomial pathogen, Vancomycin-resistant enterococci, VRE bacteremia.
INTRODUCTION
Enterococci are part of the normal resident flora of the gastrointestinal tract of humans and animals.1 It is implicated to cause various community and hospital-acquired infections, such as endocarditis, bacteremia, meningitis, and urinary tract infections, and is also associated with intra-abdominal infections.1 Recently, enterococci are emerging as an important multidrug-resistant pathogen causing nosocomial infections, predominantly bacteremia and urinary tract infections.2 Particularly, glycopeptide resistance in enterococci leads to an increased length of hospital stay and mortality, hence causing high public health threats due to limited treatment options. In 1988, the first case of vancomycin-resistant enterococci (VRE) was reported in Europe.3 In India, the first VRE case was reported in New Delhi in 1999, and the prevalence reports of VRE in India vary from 1 to 8.7%.4 The World Health Organization recognized VRE as one of the most significant resistant bacteria in their “Global Priority list of antibiotic-resistant bacteria” in 2017.5 The Centers for Disease Control and Prevention reported that VRE has caused 54,500 infections among hospitalized patients and 5,400 estimated deaths in the United States in 2017.6 The European Antimicrobial Resistance Surveillance Network reported the increasing trend of the mean proportion of vancomycin-resistant Enterococcus faecium in invasive isolates from 10.4% in 2014 to 17.3% in 2018 in countries of the European Union and European Economic Area.7 Enterococcus faecalis and E. faecium are the common species causing clinical infection accounting for around 80–90% and 5–10%, respectively.8 The widespread use of vancomycin and extended-spectrum cephalosporins in hospitals leads to the worldwide emergence of VRE.9 Though the rate of isolation of VRE is currently not very high in India when compared to other countries (USA and Europe), still it is definitely on an increasing side.10 It was found that the mortality and morbidity are more with VRE bacteremia.11 Therefore, in this study, we analyzed the current trends of VRE bacteremia, the clinical, microbiological, and epidemiological features of the VRE culture-positive patients in a tertiary care center in South India.
MATERIALS AND METHODS
This is an observational study conducted in the Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, from October 2017 to September 2020. All consecutive patients with culture-proven bloodstream infection with Enterococcus species for the first time during the study period were included in the study. Blood culture samples from patients were sent to the microbiology laboratory by treating clinicians if clinically indicated in automated BACT/ALERT aerobic blood culture bottles. The bottles were loaded in the automated VIRTUO BACT/ALERT 3D system immediately. Once the bottle flagged a positive signal, the time to positivity (TTP) was noted. Grams staining directly from the flagged bottle were performed, followed by culture on 5% sheep blood agar and MacConkey agar along with direct antimicrobial susceptibility testing (AST) on Mueller Hinton agar from flagged bottles by Kirby Bauer disk diffusion method, and incubated aerobically at 37°C. As soon as visible colonies appear on the culture plate, the identification of the organism from the colony was performed using the matrix-assisted laser desorption ionizationtime-of-flight mass spectrometry (VITEK MS, Biomeriuex). Then, AST was performed from the isolated colony by Kirby Bauer disk diffusion method and minimum inhibitory concentration determination by automated VITEK-2 platform. The susceptibility results were interpreted after 16 to 18 hours of incubation using Clinical and Laboratory Standards Institute 2020M-100 clinical breakpoints.12 The antibiotics tested were ampicillin, high-level gentamicin, vancomycin, linezolid, and teicoplanin. Susceptibility for vancomycin was also screened by performing spot inoculation from culture inoculum on vancomycin screen agar plate (6 µg/mL) to identify VRE isolates. The quality control strains used for vancomycin screening were Staphylococcus aureus ATCC 25923 and E. faecalis ATCC 29212. The demographic details, clinical profile, and the clinical outcome of the patients with VRE bacteremia were collected and analyzed.
RESULTS
A total of 81,437 blood culture samples were received from the microbiology laboratory during the study period (October 2017–September 2020), among which 15,922 bottles flagged positive. Out of these positive bottles, after excluding the duplicate isolates, 427 grew Enterococcus species. Of these, 195 (45.6%) were E. faecium and 186 (43.5%) were E. faecalis. Other species of Enterococcus isolated were E. avium, E. hirae, E. casseliflavis, and E. gallinarum, and no species were identified in 28 isolates of enterococci (Table 1). Of the total 427 Enterococcus species isolated, 63 (14.7%) were found to be resistant to vancomycin with the yearly vancomycin resistant rate of 6.12% (2018), 13.2% (2019), and 19.2% (2020) (Fig. 1). The distribution of VRE cases based on the hospital location was shown in Figures 2 and 3. About 43 patients (68%) were located in intensive care units (ICUs), and the rest 20 patients (32%) were from non-ICU location like inpatient wards. For a total of 16 patients with VRE bacteremia (25.3%), clinical diagnosis was mentioned as sepsis. A total of nine patients with VRE bacteremia (14.2%) had chronic kidney disease (CKD) as comorbidity (Table 2). The average TTP of VRE isolates was found to be 13.2 hours, and there is no significant difference in TTP between the vancomycin-resistant and susceptible isolates. VanA gene (81%)-mediated resistance was found to be more common followed by VanC (11%) and VanB (8%) (Fig. 4). The susceptibility pattern of the VRE isolates to other commonly used antibiotics showed high susceptibility to linezolid [50/63 (79.3%)] and high-level gentamicin [45/63 (71.4%)] and reduced susceptibility to ampicillin [18/63 (28.5%)], levofloxacin [21/63 (33.3%)], and teicoplanin [5/63 (7.9%)] (Fig. 5).
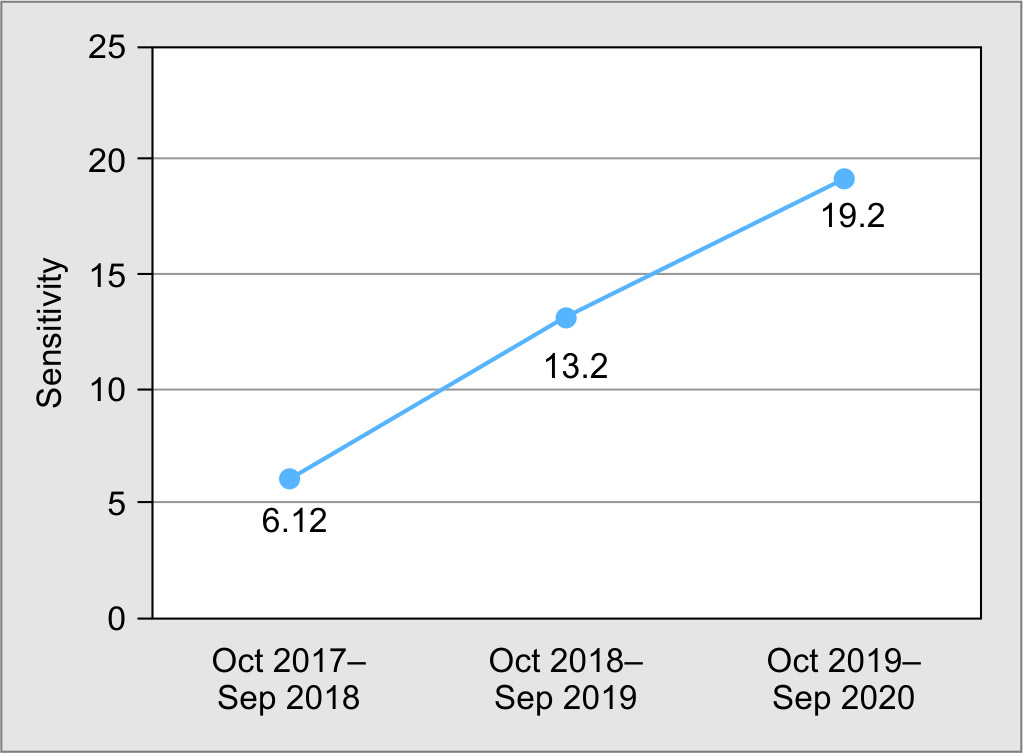
Fig. 1: Vancomycin-resistant enterococci rate from October 2017 to September 2020

Fig. 2: Distribution of vancomycin-resistant enterococci

Fig. 3: Distribution of vancomycin-resistant enterococci in intensive care units. EMS ICU, emergency service intensive care unit; SICU, surgical intensive care unit; CCU, critical care unit; MICU, medicine intensive care unit; PICU, pediatrics intensive care unit

Fig. 4: Type of vancomycin-resistant enterococci isolated

Fig. 5: Susceptibility of vancomycin-resistant enterococci to other antibiotics
| Enterococcus species in blood culture | October 2017–September 2018 | October 2018–September 2019 | October 2019–September 2020 |
|---|---|---|---|
| Enterococcus faecium | 17 (17.3%) | 51 (48%) | 127 (57%) |
| Enterococcus faecalis | 66 (67.3%) | 37 (35%) | 83 (37%) |
| Enterococcus avium | 0 | 0 | 3 (1.3%) |
| Enterococcus hirae | 0 | 2 (1.8%) | 3 (1.3%) |
| Enterococcus casseliflavis | 1 (1%) | 4 (3.7%) | 0 |
| Enterococcus gallinarum | 0 | 1 (0.9%) | 4 (1.7%) |
| Enterococcus species (unidentified) | 14 (14.2%) | 11 (10.3%) | 3 (1.3%) |
| Total | 98 | 106 | 223 |
| Clinical diagnosis | No. of VRE isolated(%) |
|---|---|
| Sepsis | 16 (25.3%) |
| Chronic kidney disease | 9 (14.28%) |
| GI infections (intestinal perforations, gut gangrene, post-ileostomy, etc.) | 7 (11.1%) |
| Acute myeloid leukemia | 8 (12.6%) |
| Pleural effusion/severe acute respiratory illness | 5 (7.9%) |
| Infective endocarditis | 1 (1.5%) |
| Diabetic foot/diabetic keto acidosis | 2 (3.17%) |
| Others | 16 (25.3%) |
| Total | 63 |
DISCUSSION
The importance of Enterococcal infections is increasing not only because of the seriousness of the infections caused but also owing to the development of resistance to various antibiotics. VRE has emerged as a leading cause of nosocomial infections, and VRE bacteremia is an independent risk factor for mortality.13 Glycopeptide resistance was found to be mediated by different vancomycin resistance (Van) gene operons, such as VanA, VanB, VanC (C1, C2, and C3), VanD, VanE, VanG, VanL, VanM, and VanN.14 Among these, resistance mediated by VanA is most common followed by VanB. VanC is responsible for the intrinsic resistance seen in E. gallinarum and E. casseliflavus.15 Enterococci have also been identified to transfer its vancomycin-resistant gene clusters to methicillin-resistant S. aureus by horizontal gene transfer, which is considered as a public threat.16 Uncontrolled and excessive use of cell wall-acting antibiotics, including vancomycin, leading to antibiotic selective pressure is the single most important predisposing factor responsible for the emergence of resistance to vancomycin.8,17
In India, the prevalence of VRE infections is in the increasing trend varying from 1 to 8.7%.4,18 Our study also shows that VRE infection rate in bloodstream is in the increasing trend from 6.12% in 2018 to 19.2% in 2020. Another study by Deshpande et al. done in a tertiary care center in Mumbai also has reported the prevalence of VRE to be 19.6%.19 Data obtained from other countries like Germany also revealed the increase in VRE rate from less than 5% in 2001 to 14.5% in 2013, mainly vancomycin-resistant E. faecium.20 The most common species of Enterococcus isolated during the study period is E. faecium, although it varied between years. Initially, from 2017 to 2018, E. faecalis was the predominant isolate, but gradually, the frequency of isolation of E. faecium, which is more drug-resistant, has started increasing. This is in concordance with the Indian study conducted by Khandelwal et al. in April 2020 in a tertiary care center in Gujarat, where E. faecium is the predominant species isolated.18 In contrast, another Indian study conducted by Deshpande et al. in a tertiary care center in Mumbai has shown that E. faecalis to be the commonly isolated species than E. faecium.19 A study on VRE bacteremia surveillance conducted in Switzerland has also shown that E. faecalis to be the commonest species isolated.
In the present study, middle-aged and elderly adult males [50/63 (79.3%)] with the mean age of 56 years are the predominant age-group from whom VRE has been isolated. Various other studies also have shown that the VRE colonization and infection being common between the age of 55 years and 75 years.11,21 Common risk factors for VRE colonization and bacteremia include immunosuppression, neutropenia, renal insufficiency, prolonged hospital stay, ICU admission, staying in close proximity to a VRE patient, and extensive antimicrobial use.22,23
In our study, prolonged ICU stay due to sepsis (25.3%), CKD (14.28%), acute myeloid leukemia (12.6%), and gastrointestinal (GI) infections and surgeries (11.1%) are the common risk factors associated with VRE bacteremia. The majority of the VRE cases [43/63 (68%)] were isolated from the patients admitted to ICUs than from non-ICU locations. In concordance, several other studies have also shown that prolonged ICU stay to be one of the most important causes for a significant increase in the VRE rate.24–27
In our study, the most common VRE phenotype detected is VanA (81%), and it produces inducible, high-level resistance to vancomycin (MICs, ≥64 µg/mL) and teicoplanin (MICs, ≥16 µg/ mL).17 This occurs due to the substitution of D-Ala-D-Ala peptide on NAM subunits with D-Ala-D-Lac, which reduces the affinity of vancomycin for the pentapeptide to 1,000-folds.15 VanB-mediated resistance is comparatively rare (8%), and it confers moderate- to high-level resistance to vancomycin (varied from 4 to 256 µg/mL) but remains susceptible to teicoplanin.15 VanC-mediated resistance attributed to 11% of total VRE, which confers intrinsic resistance to both vancomycin and teicoplanin.17 This finding is in concordance with other studies where VanA is the most common phenotype to be isolated.2,28
The susceptibility pattern of the VRE isolates to other commonly used antibiotics was found to be variable. The majority of the isolates were susceptible to linezolid (79.3%) and high-level gentamicin (71.4%), whereas reduced susceptibility was observed in antibiotics like levofloxacin (33.3%), ampicillin (28.5%), and teicoplanin (7.9%). Among the VRE isolates, 8/63 (12.7%) were found to be resistant to linezolid. Linezolid-resistant enterococci and VRE are also an emerging global threat as there is limited treatment option available.19 The above data show that there is a high prevalence of multidrug-resistant enterococci in our setting. This concomitant high-level resistance to other antibiotics will have adverse therapeutic and clinical implications, as it narrows down the therapeutic options available for treating enterococcal infections.
The present study has few limitations. Only blood isolates were included in this study. The VRE distribution in other specimens was not studied. This study did not involve the screening for VRE colonization of the other patients in ICU and wards where VRE has been isolated from clinical specimens. The clinical outcome, follow-up, and infection control measures taken for the patients with VRE bacteremia were not studied.
CONCLUSION
In this study, we found a rapid surge of VRE bacteremia in our healthcare setting, which is probably due to the overuse of antibiotics and prolonged hospital stay. It is very important to identify, treat, and take preventive measures to limit the spread of VRE, which otherwise will result in serious consequences. Active screening of at-risk patients for VRE colonization is an important step to initiate appropriate infection control measures that include isolation and cohort of the VRE colonized patients, using patient-dedicated equipment, absolute adherence to hand hygiene, and cleaning the room effectively after the discharge of the patient. It is also important to follow principles of antimicrobial stewardship, such as avoidance of use of vancomycin for surgical antimicrobial prophylaxis, avoiding the empirical use of vancomycin if not indicated, and de-escalating from vancomycin to a susceptible narrow-spectrum antibiotic when the blood culture is negative for beta-lactam-resistant Gram-positive microorganisms.15,29 This increasing trend of VRE bacteremia is a red alert to the clinicians, and strict antibiotic policies and proper adherence to the infection control practices should be followed to reduce the VRE rate.
ORCID
Monika Sivaradjy https://orcid.org/0000-0002-3193-4258
Anitha Gunalan https://orcid.org/0000-0002-7779-7877
Ketan Priyadarshi https://orcid.org/0000-0003-4623-3523
Haritha Madigubba https://orcid.org/0000-0001-5486-0821
Deepashree Rajshekar https://orcid.org/0000-0001-9124-1173
Apurba S Sastry https://orcid.org/0000-0003-2337-3830
REFERENCES
1. Bourdon N, Fines-Guyon M, Thiolet JM, Maugat S, Coignard B, Leclercq R, et al. Changing trends in vancomycin-resistant enterococci in French hospitals, 2001–08. J Antimicrob Chemother 2011;66(4):713–721. DOI: 10.1093/jac/dkq524.
2. Praharaj I, Sujatha S, Parija SC. Phenotypic and genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res 2013;138(4):549–556. PMID: 24434263.
3. Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis 2006;42(Suppl. 1):S25–S34. DOI: 10.1086/491711.
4. Purohit G, Gaind R, Dawar R, Verma PK, Aggarwal KC, Sardana R, et al. Characterization of Vancomycin Resistant Enterococci in hospitalized patients and role of gut colonization. J Clin Diagn Res 2017;11:5. DOI: 10.7860/JCDR/2017/25988.10548.
5. Tacconelli E, Magrini N, Kahlmeter G, Singh N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization 2017;27:318–327. DOI: 10.1016/S1473-3099(17)30753-3.
6. Centers for Disease Control and Prevention. Vancomycin-resistant enterococci (VRE) in healthcare settings.
7. Ayobami O, Willrich N, Reuss A, Eckmanns T, Markwart R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of bloodstream infections. Emerg Microbes Infect 2020;9(1):1180–1193. DOI: 10.1080/22221751.2020.1769500.
8. Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev 2000;13:22. DOI: 10.1128/cmr.13.4.686-707.2000.
9. Rice LB. Emergence of vancomycin-resistant enterococci. Emerg Infect Dis 2001;7(2):183–187. DOI: 10.3201/eid0702.010205.
10. Vidyalakshmi P, Ramasubramanian V, Nambi Ps, Gopalakrishnan R, Ghafur Ka, Thirunarayana M. Clinical, epidemiological, and microbiological profile of patients with vancomycin-resistant Enterococci from a Tertiary Care Hospital. J Global Infect Dis 2012;4(2):137. DOI: 10.4103/0974-777X.96784.
11. Piezzi V, Gasser M, Atkinson A, Kronenberg A, Vuichard-Gysin D, Harbarth S, et al. Increasing proportion of vancomycin resistance among enterococcal bacteremias in Switzerland: a 6-year nation-wide surveillance, 2013 to 2018. Euro Surveill 2020;25(35):1900575. DOI: 10.2807/1560-7917.ES.2020.25.35.1900575.
12. CLSI, Clinical and Laboratory Standard Institute. Clinical and Laboratory Standards Institute CLSI document: performance standard for antimicrobial susceptibility testing; informational supplement M100-S21.
13. Montecalvo MA, Shay DK, Patel P, Tacsa L, Maloney SA, Jarvis WR, et al. Bloodstream infections with vancomycin-resistant enterococci. Arch Intern Med 1996;156(13):1458–1462. DOI:10.1128/cmr.13.4.686-707.2000
14. Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, et al. vanM, A new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 2010;54(11):4643–4647. DOI: 10.1128/AAC.01710-09.
15. Faron ML, Ledeboer NA, Buchan BW. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant enterococcus in the health care setting. J Clin Microbiol 2016;54(10):2436–2447. DOI: 10.1128/JCM.00211-16.
16. Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti-infect Ther 2014;12(10):1221–1236. DOI: 10.1586/14787210.2014.956092.
17. Gold HS. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis 2001;33(2):210–219. DOI: 10.1086/321815.
18. Khandelwal N, Panwala T, Patel JS. Prevalence of enterococcus species and its vancomycin resistance pattern in a Tertiary Care Hospital, Surat, Gujarat, India: a growing threat. Int J Recent Sci Res 2020;11(7):3. DOI: 10.24327/ijrsr.2020.1107.5480.
19. Deshpande VR, Karmarkar MG, Mehta PR. Prevalence of multidrug-resistant enterococci in a tertiary care hospital in Mumbai, India. J Infect Dev Ctries 2013;7(02):155–158. DOI: 10.3855/jidc.3018.
20. Gastmeier P, Schroder C, Behnke M, Meyer E, Geffers C. Dramatic increase in vancomycin-resistant enterococci in Germany. J Antimicrob Chemother 2014;69(6):1660–1664. DOI: 10.1093/jac/dku035.
21. Sohn KM. Duration of colonization and risk factors for prolonged carriage of vancomycin-resistant enterococci after discharge from the hospital. Int J Infect Dis 2013;17(4):e240–e246. DOI: 10.1016/j.ijid.2012.09.019.
22. Mathis B, Haïne M, Girard R, Bonnefoy M. Risk factors for vancomycin-resistant enterococcus acquisition during a large outbreak in patients aged 65 years and older. BMC Geriatr 2019;19(1):377. DOI: 10.1186/s12877-019-1398-2.
23. Zaas AK, Song X, Tucker P, Perl TM. Risk factors for development of vancomycin-resistant enterococcal bloodstream infection in patients with cancer who are colonized with vancomycin-resistant enterococci. Clin Infect Dis 2002;35(10):1139–1146. DOI: 10.1086/342904.
24. Fridkin SK, Edwards JR, Courval JM, Hill H, Tenover FC, Lawton R, et al. The effect of vancomycin and third-generation cephalosporins on prevalence of vancomycin-resistant enterococci in 126 U.S. adult intensive care units. Ann Intern Med 2001;135(3):175–183. DOI: 10.7326/0003-4819-135-3-200108070-00009.
25. Moemen D, Tawfeek D, Badawy W. Healthcare-associated vancomycin resistant Enterococcus faecium infections in the Mansoura University Hospitals intensive care units, Egypt. Braz J Microbiol 2015;46(3):777–783. DOI: 10.1590/S1517-838246320140403.
26. Monteserin N, Larson E. Temporal trends and risk factors for healthcare-associated vancomycin-resistant enterococci in adults. J Hosp Infect 2016;94(3):236–241. DOI: 10.1016/j.jhin.2016.07.023.
27. Remschmidt C, Schröder C, Behnke M, Gastmeier P, Geffers C, Kramer TS. Continuous increase of vancomycin resistance in enterococci causing nosocomial infections in Germany − 10 years of surveillance. Antimicrob Resist Infect Control 2018;7(1):54. DOI: 10.1186/s13756-018-0353-x.
28. Moosavian M, Ghadri H, Samli Z. Molecular detection of vanA and vanB genes among vancomycin-resistant enterococci in ICU-hospitalized patients in Ahvaz in southwest of Iran. Infect Drug Resist 2018;11:2269–2275. DOI: 10.2147/IDR.S177886.
29. Recommendations for preventing the spread of vancomycin resistance: recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). Am J Infect Control 1995;23(2):87–94. DOI: 10.1016/0196-6553(95)90104-3.
________________________
© The Author(s). 2021 Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and non-commercial reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.